2026
Rethinking the Trust Region in LLM Reinforcement Learning
Penghui Qi*, Xiangxin Zhou*, Zichen Liu, Tianyu Pang, Chao Du, Min Lin, Wee Sun Lee (* equal contribution)
Preprint. 2026
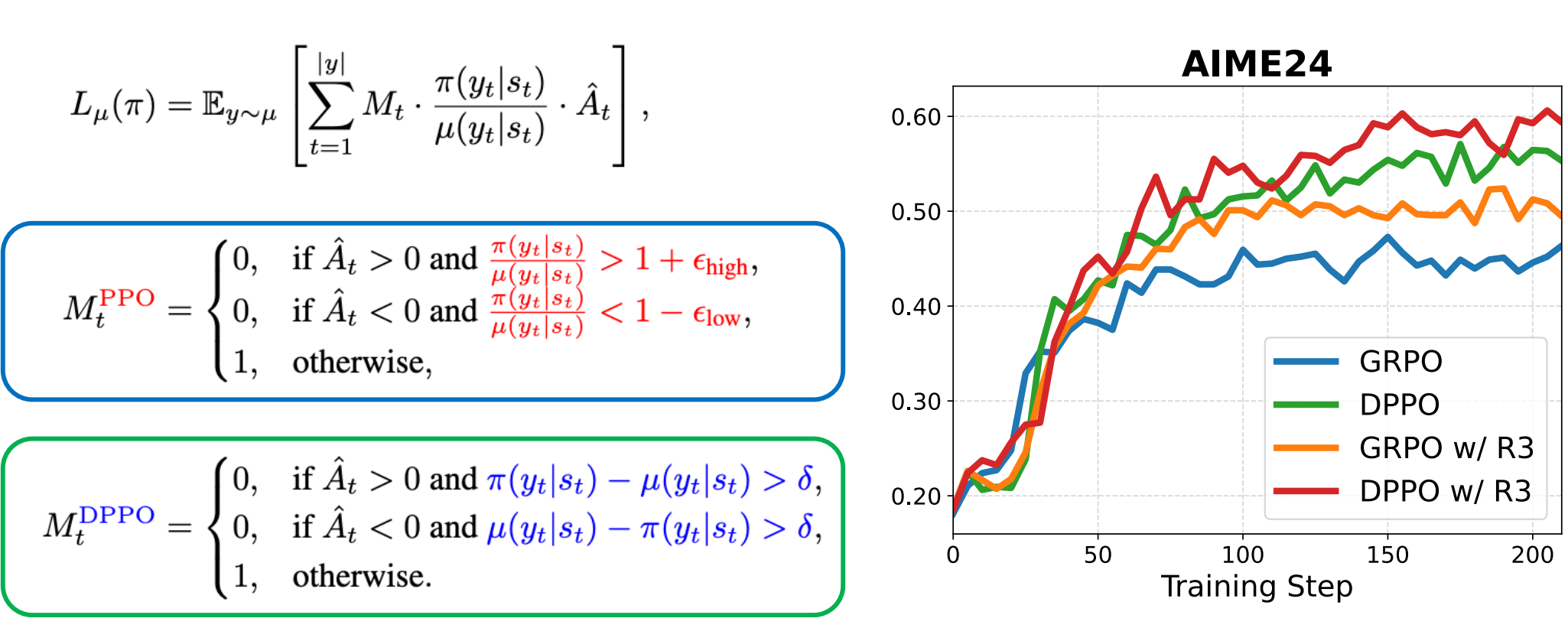
We argue that the core ratio clipping mechanism in PPO is structurally ill-suited for the large vocabularies inherent to LLMs. PPO constrains policy updates based on the probability ratio of sampled tokens, which serves as a noisy single-sample Monte Carlo estimate of the true policy divergence. This motivate us to propose Divergence Proximal Policy Optimization (DPPO). Extensive experiments show its superior training stability and efficiency, offering a more robust foundation for LLM RL.
Rethinking the Trust Region in LLM Reinforcement Learning
Penghui Qi*, Xiangxin Zhou*, Zichen Liu, Tianyu Pang, Chao Du, Min Lin, Wee Sun Lee (* equal contribution)
Preprint. 2026
We argue that the core ratio clipping mechanism in PPO is structurally ill-suited for the large vocabularies inherent to LLMs. PPO constrains policy updates based on the probability ratio of sampled tokens, which serves as a noisy single-sample Monte Carlo estimate of the true policy divergence. This motivate us to propose Divergence Proximal Policy Optimization (DPPO). Extensive experiments show its superior training stability and efficiency, offering a more robust foundation for LLM RL.
Variational Reasoning for Language Models
Xiangxin Zhou*, Zichen Liu, Haonan Wang, Chao Du, Min Lin, Chongxuan Li, Liang Wang, Tianyu Pang* (* equal contribution)
International Conference on Learning Representations (ICLR) 2026
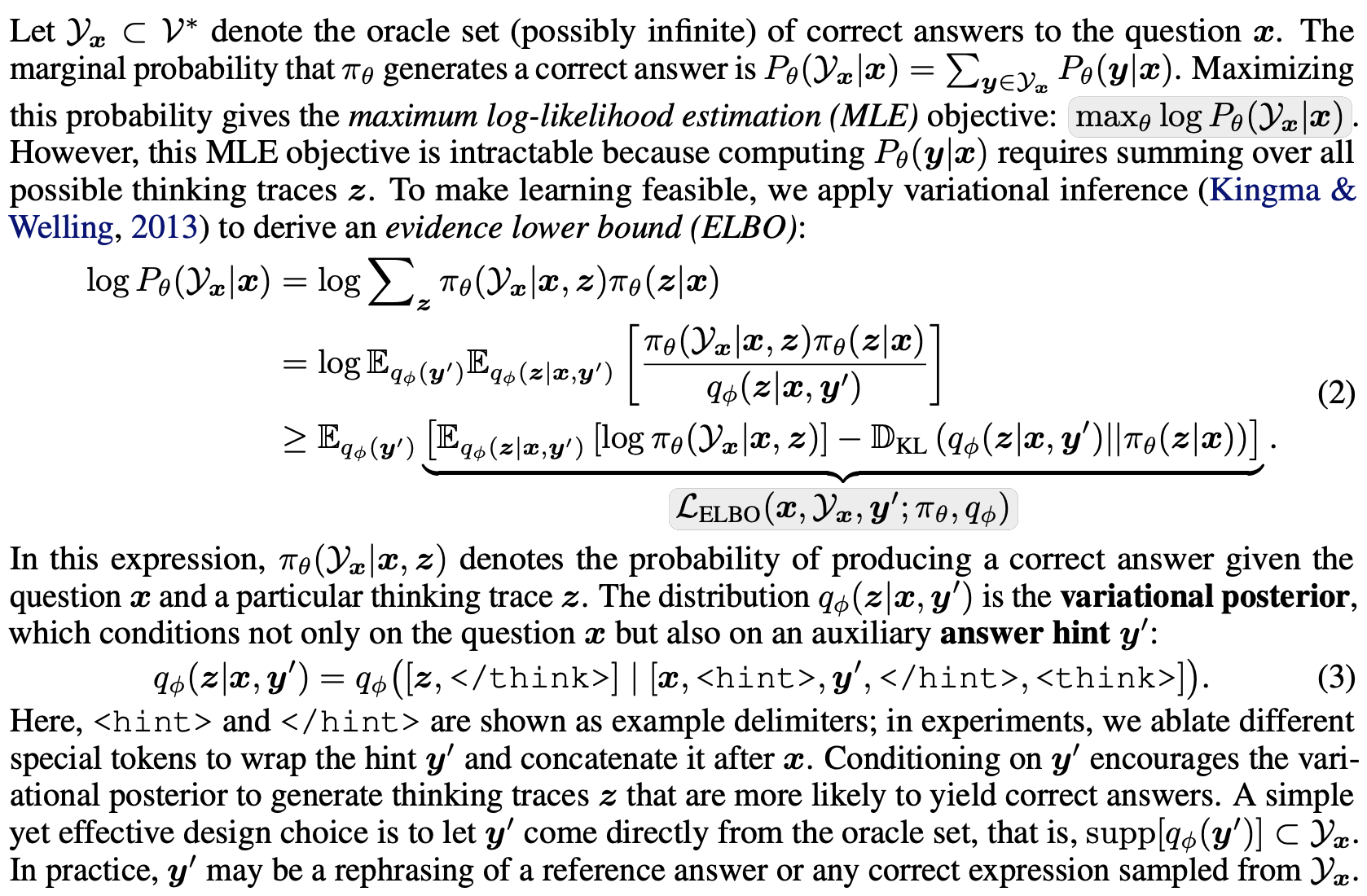
We introduce a variational reasoning framework for language models that treats thinking traces as latent variables and optimizes them through variational inference. Starting from the evidence lower bound (ELBO), we extend it to a multi-trace objective for tighter bounds and propose a forward-KL formulation that stabilizes the training of the variational posterior. This framework provides a principled probabilistic perspective that unifies variational inference with RL-style methods and yields stable objectives for improving the reasoning ability of language models.
Variational Reasoning for Language Models
Xiangxin Zhou*, Zichen Liu, Haonan Wang, Chao Du, Min Lin, Chongxuan Li, Liang Wang, Tianyu Pang* (* equal contribution)
International Conference on Learning Representations (ICLR) 2026
We introduce a variational reasoning framework for language models that treats thinking traces as latent variables and optimizes them through variational inference. Starting from the evidence lower bound (ELBO), we extend it to a multi-trace objective for tighter bounds and propose a forward-KL formulation that stabilizes the training of the variational posterior. This framework provides a principled probabilistic perspective that unifies variational inference with RL-style methods and yields stable objectives for improving the reasoning ability of language models.
Reinforcing General Reasoning Without Verifiers
Xiangxin Zhou*, Zichen Liu*, Anya Sims*, Haonan Wang, Tianyu Pang, Chongxuan Li, Liang Wang, Min Lin, Chao Du (* equal contribution)
International Conference on Learning Representations (ICLR) 2026
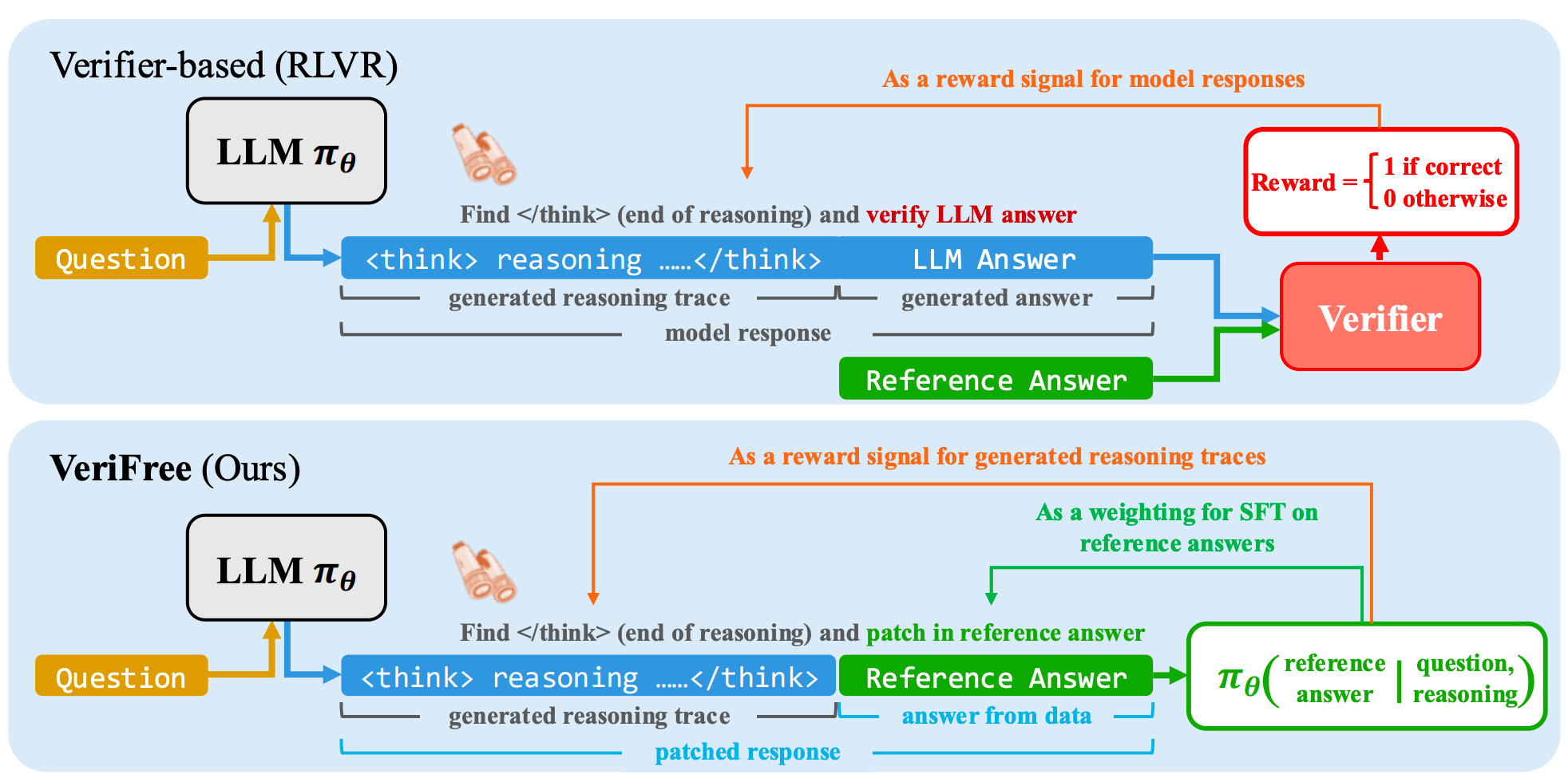
VeriFree is a verifier-free method that bypasses answer verification and instead uses RL to directly maximize the probability of generating the reference answer.
Reinforcing General Reasoning Without Verifiers
Xiangxin Zhou*, Zichen Liu*, Anya Sims*, Haonan Wang, Tianyu Pang, Chongxuan Li, Liang Wang, Min Lin, Chao Du (* equal contribution)
International Conference on Learning Representations (ICLR) 2026
VeriFree is a verifier-free method that bypasses answer verification and instead uses RL to directly maximize the probability of generating the reference answer.

GEM: A Gym for Agentic LLMs
Zichen Liu*, Anya Sims*, Keyu Duan*, Changyu Chen*, Simon Yu, Xiangxin Zhou, Haotian Xu, Shaopan Xiong, Bo Liu, Chenmien Tan, Chuen Yang Beh, Weixun Wang, Hao Zhu, Weiyan Shi, Diyi Yang, Michael Shieh, Yee Whye Teh, Wee Sun Lee, Min Lin (* equal contribution)
International Conference on Learning Representations (ICLR) 2026
To facilitate this transition we introduce GEM (General Experience Maker), an open-source environment simulator designed for the age of LLMs. Analogous to OpenAI-Gym for traditional reinforcement learning (RL), GEM provides a standardized framework for the environment-agent interface, including asynchronous vectorized execution for high throughput, and flexible wrappers for easy extensibility. GEM also features a diverse suite of environments, robust integrated tools, and single-file example scripts demonstrating using GEM with five popular RL training frameworks.
GEM: A Gym for Agentic LLMs
Zichen Liu*, Anya Sims*, Keyu Duan*, Changyu Chen*, Simon Yu, Xiangxin Zhou, Haotian Xu, Shaopan Xiong, Bo Liu, Chenmien Tan, Chuen Yang Beh, Weixun Wang, Hao Zhu, Weiyan Shi, Diyi Yang, Michael Shieh, Yee Whye Teh, Wee Sun Lee, Min Lin (* equal contribution)
International Conference on Learning Representations (ICLR) 2026
To facilitate this transition we introduce GEM (General Experience Maker), an open-source environment simulator designed for the age of LLMs. Analogous to OpenAI-Gym for traditional reinforcement learning (RL), GEM provides a standardized framework for the environment-agent interface, including asynchronous vectorized execution for high throughput, and flexible wrappers for easy extensibility. GEM also features a diverse suite of environments, robust integrated tools, and single-file example scripts demonstrating using GEM with five popular RL training frameworks.
h-MINT: Modeling Pocket-Ligand Binding with Hierarchical Molecular Interaction Network
Yanru Qu*, Yijie Zhang*, Wenjuan Tan, Xiangzhe Kong, Xiangxin Zhou, Chaoran Cheng, Mathieu Blanchette, Jiaxuan You, Ge Liu (* equal contribution)
International Conference on Learning Representations (ICLR) 2026
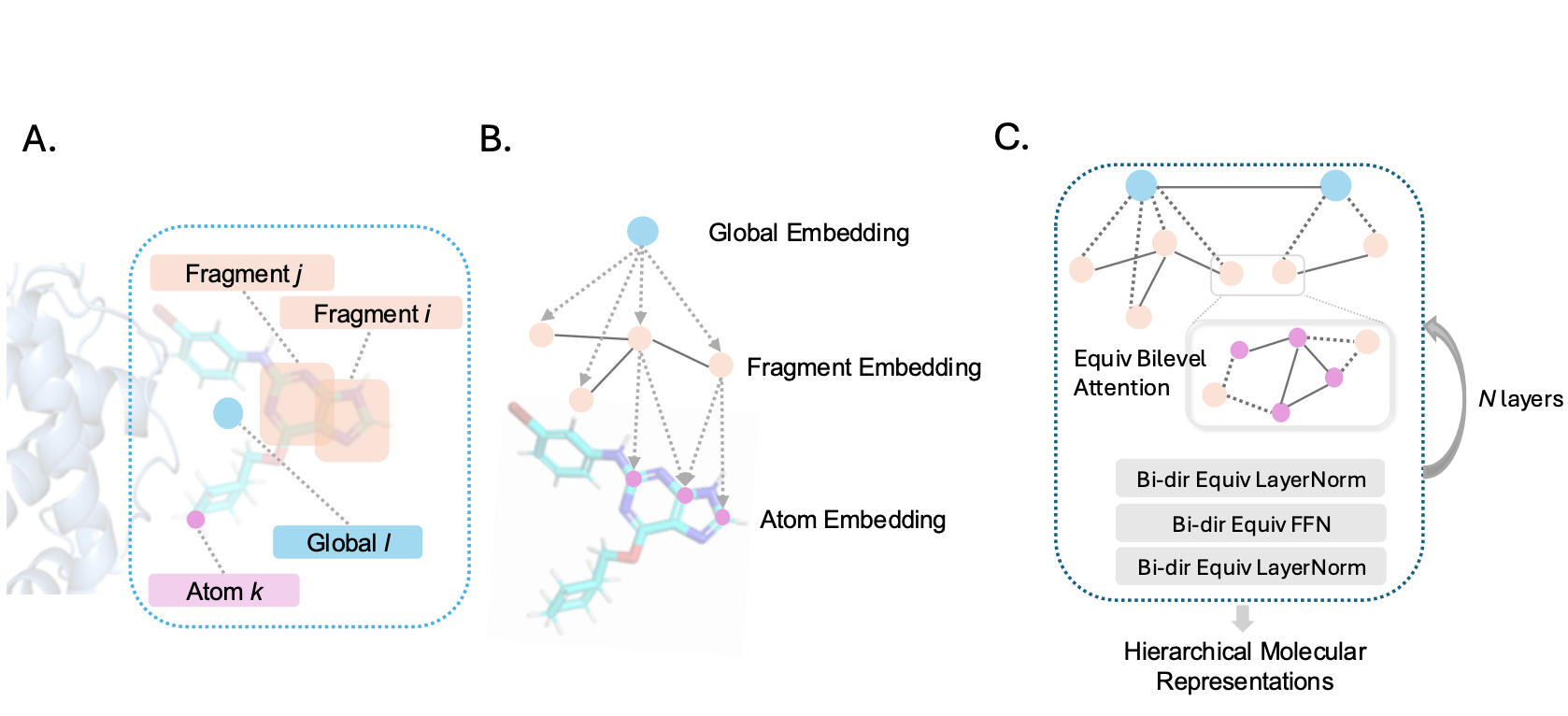
Aiming at accurate molecular representations, we propose: (i) OverlapBPE tokenization: a novel data-driven molecule tokenization that allows overlapping fragments and reflects the inherently fuzzy boundaries of small-molecule substructures; (ii) h-MINT model: a hierarchical molecular interaction network capable of jointly modeling drug–target interactions at both atom and fragment levels. Our method improves binding affinity prediction on PDBBind and LBA, enhances virtual screening on DUD-E and LIT-PCBA, and achieves the best overall HTS performance on PubChem assays.
h-MINT: Modeling Pocket-Ligand Binding with Hierarchical Molecular Interaction Network
Yanru Qu*, Yijie Zhang*, Wenjuan Tan, Xiangzhe Kong, Xiangxin Zhou, Chaoran Cheng, Mathieu Blanchette, Jiaxuan You, Ge Liu (* equal contribution)
International Conference on Learning Representations (ICLR) 2026
Aiming at accurate molecular representations, we propose: (i) OverlapBPE tokenization: a novel data-driven molecule tokenization that allows overlapping fragments and reflects the inherently fuzzy boundaries of small-molecule substructures; (ii) h-MINT model: a hierarchical molecular interaction network capable of jointly modeling drug–target interactions at both atom and fragment levels. Our method improves binding affinity prediction on PDBBind and LBA, enhances virtual screening on DUD-E and LIT-PCBA, and achieves the best overall HTS performance on PubChem assays.
2025
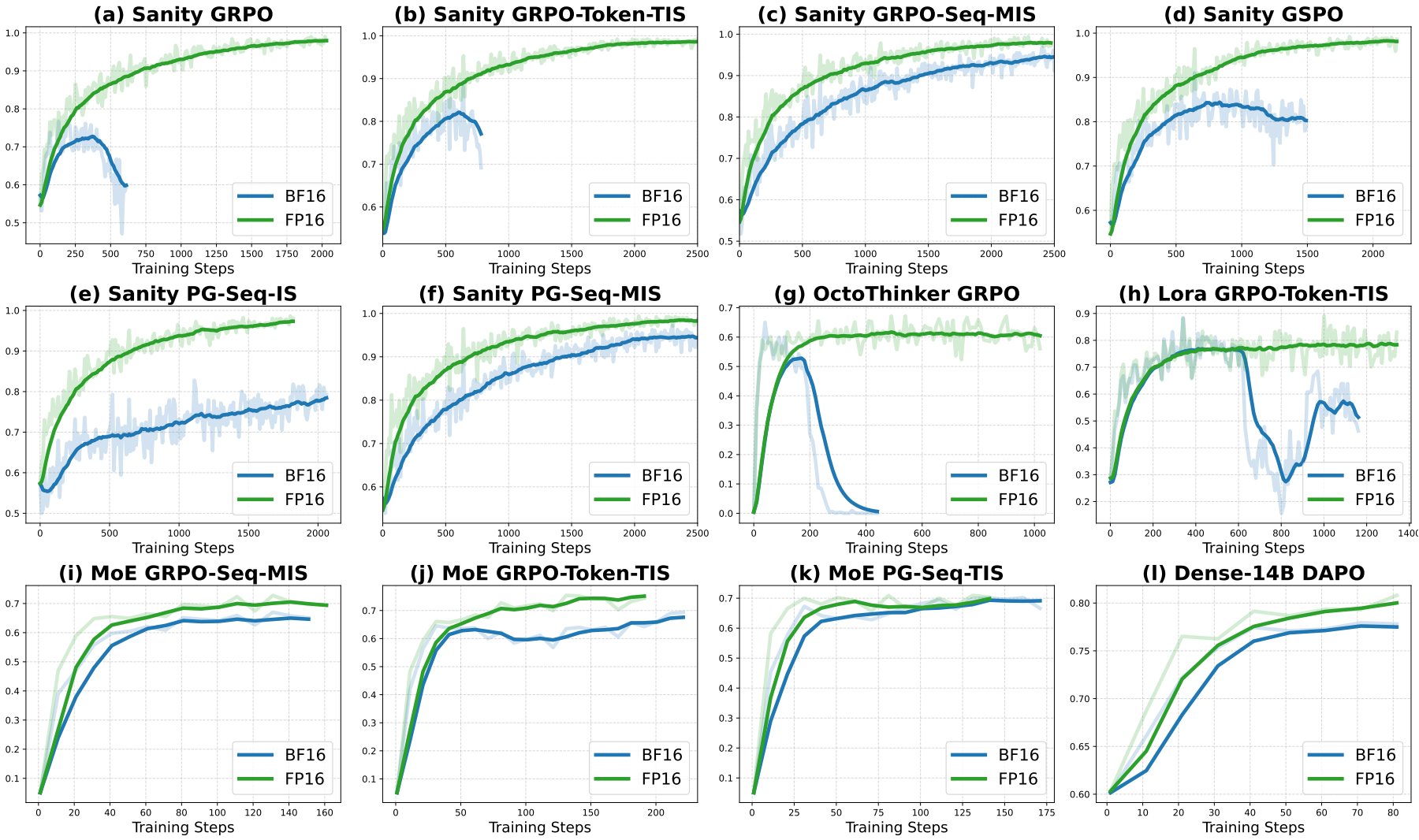
Defeating the Training-Inference Mismatch via FP16
Penghui Qi*, Zichen Liu*, Xiangxin Zhou*, Tianyu Pang, Chao Du, Wee Sun Lee, Min Lin (* equal contribution)
Preprint. 2025
We demonstrate that simply reverting to FP16 effectively eliminates the numerical mismatch between the training and inference policies in RL for LLMs. Our results suggest that using FP16 uniformly yields more stable optimization, faster convergence, and stronger performance across diverse tasks, algorithms and frameworks.
Defeating the Training-Inference Mismatch via FP16
Penghui Qi*, Zichen Liu*, Xiangxin Zhou*, Tianyu Pang, Chao Du, Wee Sun Lee, Min Lin (* equal contribution)
Preprint. 2025
We demonstrate that simply reverting to FP16 effectively eliminates the numerical mismatch between the training and inference policies in RL for LLMs. Our results suggest that using FP16 uniformly yields more stable optimization, faster convergence, and stronger performance across diverse tasks, algorithms and frameworks.
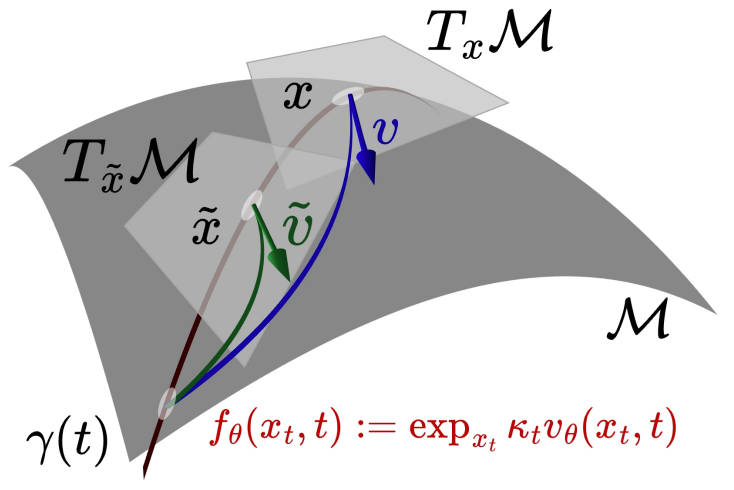
Riemannian Consistency Model
Chaoran Cheng*, Yusong Wang*, Yuxin Chen, Xiangxin Zhou, Nanning Zheng, Ge Liu (* equal contribution)
Conference on Neural Information Processing Systems (NeurIPS) 2025
We propose the Riemannian Consistency Model (RCM), which, for the first time, enables few-step consistency modeling while respecting the intrinsic manifold constraint imposed by the Riemannian geometry. Leveraging the covariant derivative and exponential-map-based parameterization, we derive the closed-form solutions for both discrete- and continuous-time training objectives for RCM.
Riemannian Consistency Model
Chaoran Cheng*, Yusong Wang*, Yuxin Chen, Xiangxin Zhou, Nanning Zheng, Ge Liu (* equal contribution)
Conference on Neural Information Processing Systems (NeurIPS) 2025
We propose the Riemannian Consistency Model (RCM), which, for the first time, enables few-step consistency modeling while respecting the intrinsic manifold constraint imposed by the Riemannian geometry. Leveraging the covariant derivative and exponential-map-based parameterization, we derive the closed-form solutions for both discrete- and continuous-time training objectives for RCM.
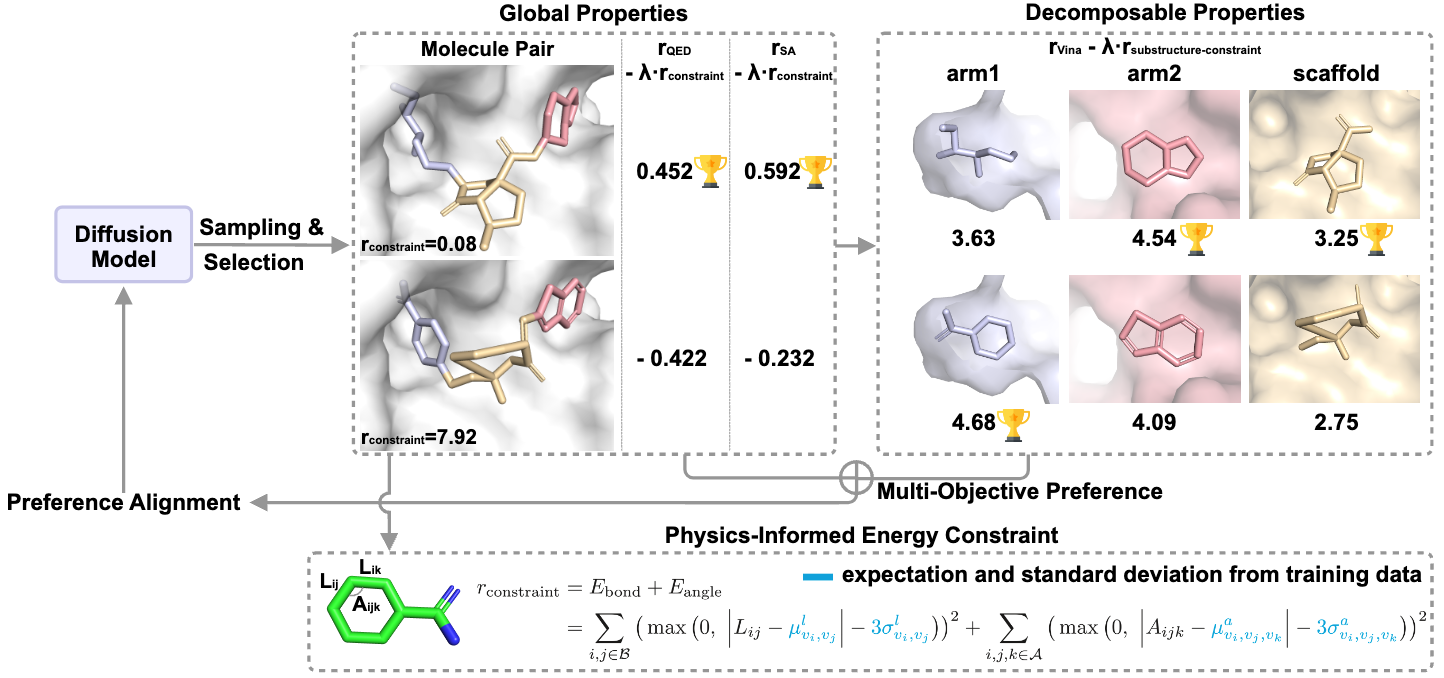
Decomposed Direct Preference Optimization for Structure-Based Drug Design
Xiwei Cheng*, Xiangxin Zhou*, Yuwei Yang, Yu Bao, Quanquan Gu (* equal contribution)
Transactions on Machine Learning Research (TMLR) 2025
We propose DecompDPO, a structure-based optimization method aligns diffusion models with pharmaceutical needs using multi-granularity preference pairs. DecompDPO introduces decomposition into the optimization objectives and obtains preference pairs at the molecule or decomposed substructure level based on each objective's decomposability. Additionally, DecompDPO introduces a physics-informed energy term to ensure reasonable molecular conformations in the optimization results.
Decomposed Direct Preference Optimization for Structure-Based Drug Design
Xiwei Cheng*, Xiangxin Zhou*, Yuwei Yang, Yu Bao, Quanquan Gu (* equal contribution)
Transactions on Machine Learning Research (TMLR) 2025
We propose DecompDPO, a structure-based optimization method aligns diffusion models with pharmaceutical needs using multi-granularity preference pairs. DecompDPO introduces decomposition into the optimization objectives and obtains preference pairs at the molecule or decomposed substructure level based on each objective's decomposability. Additionally, DecompDPO introduces a physics-informed energy term to ensure reasonable molecular conformations in the optimization results.
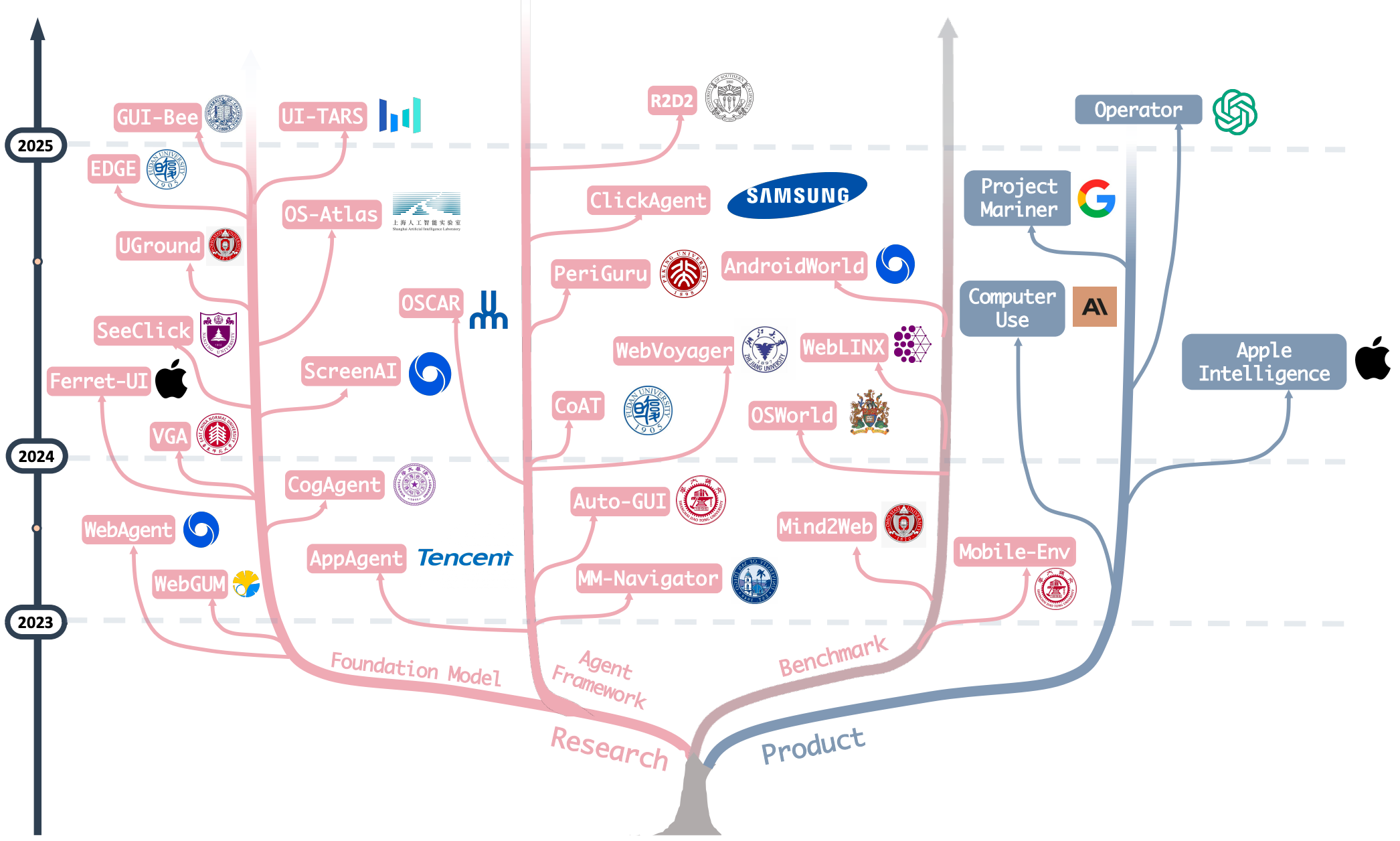
OS Agents: A Survey on MLLM-based Agents for Computer, Phone and Browser Use
Xueyu Hu, Tao Xiong, Biao Yi, Zishu Wei, Ruixuan Xiao, Yurun Chen, Jiasheng Ye, Meiling Tao, Xiangxin Zhou, Ziyu Zhao, Yuhuai Li, Shengze Xu, Shawn Wang, Xinchen Xu, Shuofei Qiao, Kun Kuang, Tieyong Zeng, Liang Wang, Jiwei Li, Yuchen Eleanor Jiang, Wangchunshu Zhou, Guoyin Wang, Keting Yin, Zhou Zhao, Hongxia Yang, Fan Wu, Shengyu Zhang, Fei Wu
Annual Meeting of the Association for Computational Linguistics (ACL) 2025 Oral
A comprehensive survey on (M)LLM-based Agents using computers, mobile phones and web browsers by operating within the environments and interfaces (e.g., Graphical User Interface (GUI) and Command Line Interface (CLI)) provided by operating systems (OS) to automate tasks.
OS Agents: A Survey on MLLM-based Agents for Computer, Phone and Browser Use
Xueyu Hu, Tao Xiong, Biao Yi, Zishu Wei, Ruixuan Xiao, Yurun Chen, Jiasheng Ye, Meiling Tao, Xiangxin Zhou, Ziyu Zhao, Yuhuai Li, Shengze Xu, Shawn Wang, Xinchen Xu, Shuofei Qiao, Kun Kuang, Tieyong Zeng, Liang Wang, Jiwei Li, Yuchen Eleanor Jiang, Wangchunshu Zhou, Guoyin Wang, Keting Yin, Zhou Zhao, Hongxia Yang, Fan Wu, Shengyu Zhang, Fei Wu
Annual Meeting of the Association for Computational Linguistics (ACL) 2025 Oral
A comprehensive survey on (M)LLM-based Agents using computers, mobile phones and web browsers by operating within the environments and interfaces (e.g., Graphical User Interface (GUI) and Command Line Interface (CLI)) provided by operating systems (OS) to automate tasks.

An All-Atom Generative Model for Designing Protein Complexes
Ruizhe Chen*, Dongyu Xue*, Xiangxin Zhou, Zaixiang Zheng, Xiangxiang Zeng, Quanquan Gu (* equal contribution)
International Conference on Machine Learning (ICML) 2025
By integrating atom-level information and leveraging data on multi-chain proteins, APM is capable of precisely modeling interchain interactions and designing protein complexes with binding capabilities from scratch.
An All-Atom Generative Model for Designing Protein Complexes
Ruizhe Chen*, Dongyu Xue*, Xiangxin Zhou, Zaixiang Zheng, Xiangxiang Zeng, Quanquan Gu (* equal contribution)
International Conference on Machine Learning (ICML) 2025
By integrating atom-level information and leveraging data on multi-chain proteins, APM is capable of precisely modeling interchain interactions and designing protein complexes with binding capabilities from scratch.

Modeling All-Atom Glycan Structures via Hierarchical Message Passing and Multi-Scale Pre-training
Minghao Xu, Jiaze Song, Keming Wu, Xiangxin Zhou, Bin Cui, Wentao Zhang
International Conference on Machine Learning (ICML) 2025
GlycanAA encodes heterogeneous all-atom glycan graphs with hierarchical message passing and is further pre-trained on unlabeled glycans through multi-scale mask prediction.
Modeling All-Atom Glycan Structures via Hierarchical Message Passing and Multi-Scale Pre-training
Minghao Xu, Jiaze Song, Keming Wu, Xiangxin Zhou, Bin Cui, Wentao Zhang
International Conference on Machine Learning (ICML) 2025
GlycanAA encodes heterogeneous all-atom glycan graphs with hierarchical message passing and is further pre-trained on unlabeled glycans through multi-scale mask prediction.
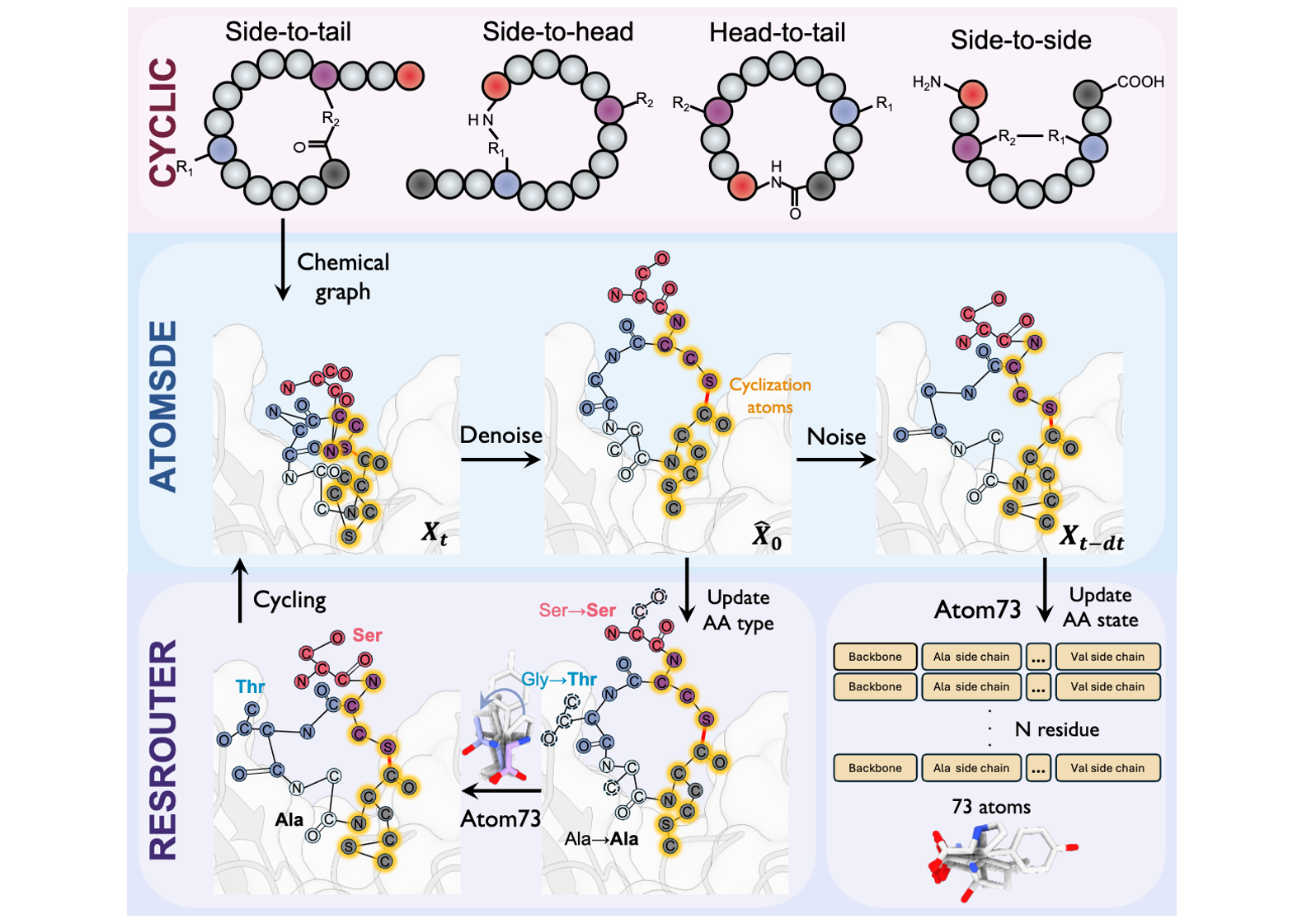
Designing Cyclic Peptides via Harmonic SDE with Atom-Bond Modeling
Xiangxin Zhou*, Mingyu Li*, Yi Xiao, Jiahan Li, Dongyu Xue, Zaixiang Zheng, Jianzhu Ma, Quanquan Gu (* equal contribution)
International Conference on Machine Learning (ICML) 2025
CpSDE is a generative algorithm capable of generating diverse types of cyclic peptides given 3D receptor structures.
Designing Cyclic Peptides via Harmonic SDE with Atom-Bond Modeling
Xiangxin Zhou*, Mingyu Li*, Yi Xiao, Jiahan Li, Dongyu Xue, Zaixiang Zheng, Jianzhu Ma, Quanquan Gu (* equal contribution)
International Conference on Machine Learning (ICML) 2025
CpSDE is a generative algorithm capable of generating diverse types of cyclic peptides given 3D receptor structures.
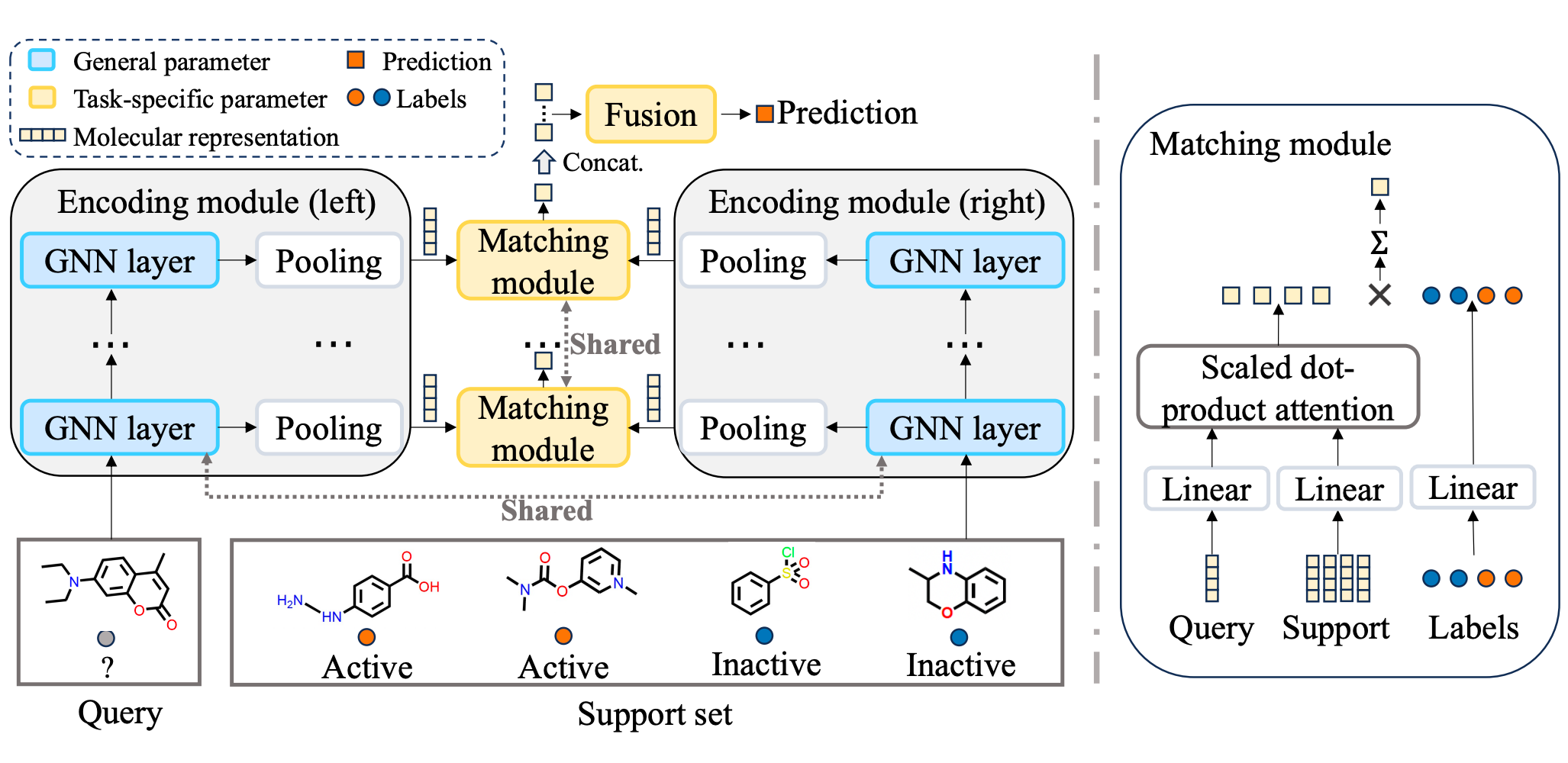
UniMatch: Universal Matching from Atom to Task for Few-Shot Drug Discovery
Ruifeng Li, Mingqian Li, Wei Liu, Yuhua Zhou, Xiangxin Zhou, Yuan Yao, Qiang Zhang, Hongyang Chen
International Conference on Learning Representations (ICLR) 2025 Spotlight
A dual matching framework that integrates explicit hierarchical molecular matching with implicit task-level matching via meta-learning, bridging multi-level molecular representations and task-level generalization.
UniMatch: Universal Matching from Atom to Task for Few-Shot Drug Discovery
Ruifeng Li, Mingqian Li, Wei Liu, Yuhua Zhou, Xiangxin Zhou, Yuan Yao, Qiang Zhang, Hongyang Chen
International Conference on Learning Representations (ICLR) 2025 Spotlight
A dual matching framework that integrates explicit hierarchical molecular matching with implicit task-level matching via meta-learning, bridging multi-level molecular representations and task-level generalization.
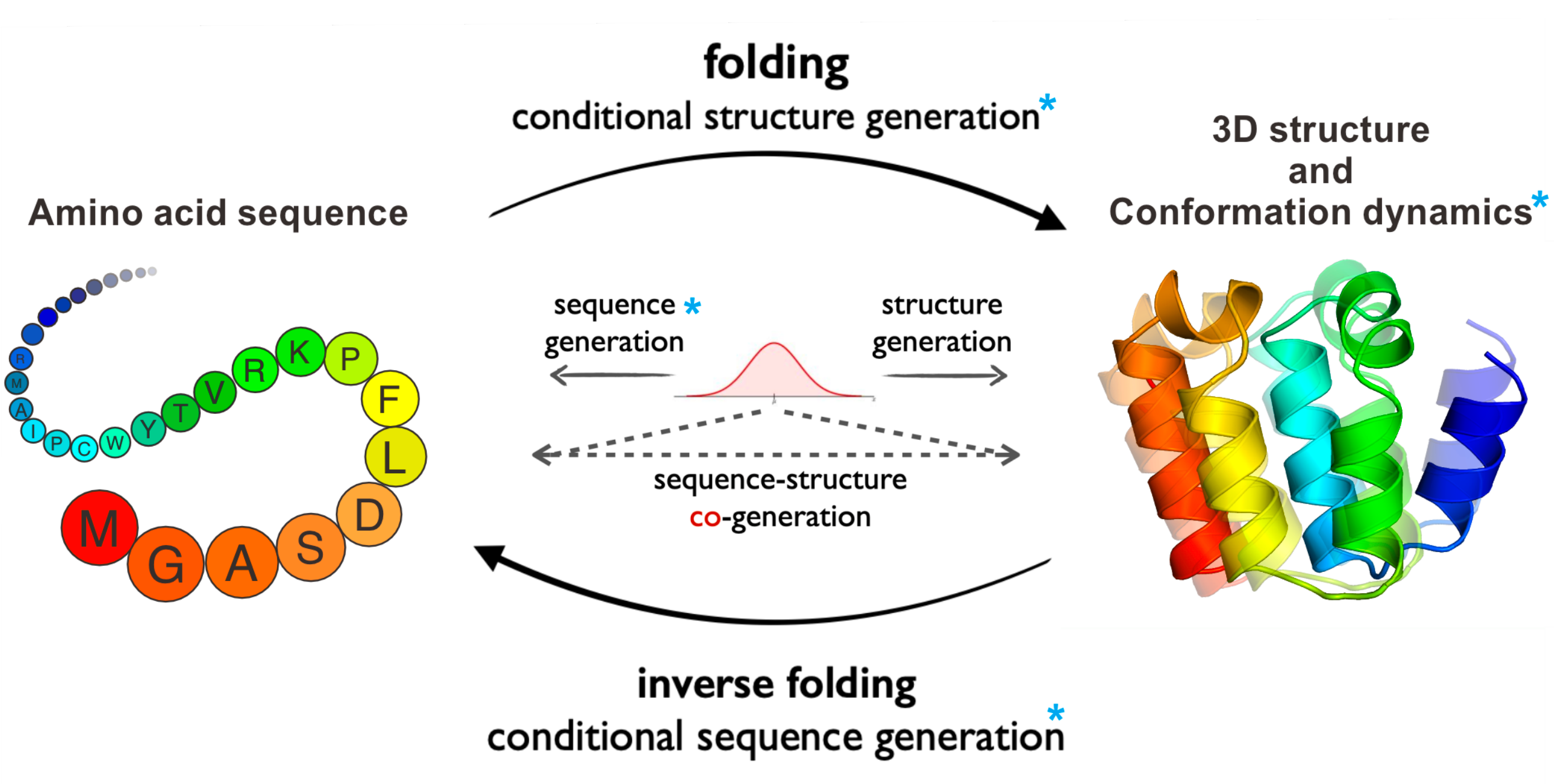
ProteinBench: A Holistic Evaluation of Protein Foundation Models
Fei Ye*, Zaixiang Zheng*, Dongyu Xue*, Yuning Shen*, Lihao Wang*, Yiming Ma, Yan Wang, Xinyou Wang, Xiangxin Zhou, Quanquan Gu (* equal contribution)
International Conference on Learning Representations (ICLR) 2025
ProteinBench is a holistic evaluation framework designed for protein prediction and generative tasks ranging from 3D structure prediction and protein design to conformational dynamics.
ProteinBench: A Holistic Evaluation of Protein Foundation Models
Fei Ye*, Zaixiang Zheng*, Dongyu Xue*, Yuning Shen*, Lihao Wang*, Yiming Ma, Yan Wang, Xinyou Wang, Xiangxin Zhou, Quanquan Gu (* equal contribution)
International Conference on Learning Representations (ICLR) 2025
ProteinBench is a holistic evaluation framework designed for protein prediction and generative tasks ranging from 3D structure prediction and protein design to conformational dynamics.
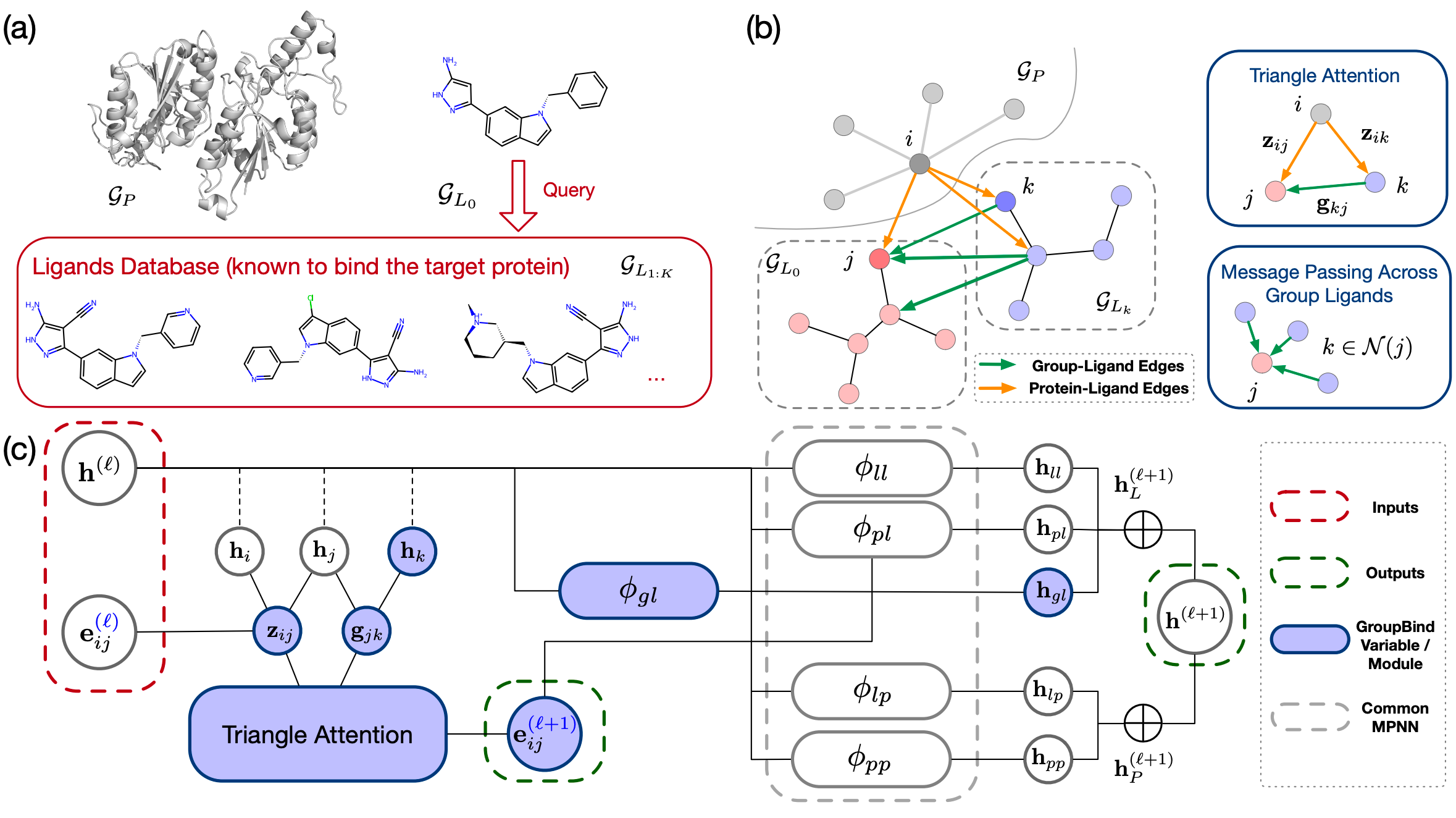
Group Ligands Docking to Protein Pockets
Jiaqi Guan*, Jiahan Li*, Xiangxin Zhou, Xingang Peng, Sheng Wang, Yunan Luo, Jian Peng, Jianzhu Ma (* equal contribution)
International Conference on Learning Representations (ICLR) 2025
GroupBind is a novel molecular docking framework that simultaneously considers multiple ligands docking to a protein.
Group Ligands Docking to Protein Pockets
Jiaqi Guan*, Jiahan Li*, Xiangxin Zhou, Xingang Peng, Sheng Wang, Yunan Luo, Jian Peng, Jianzhu Ma (* equal contribution)
International Conference on Learning Representations (ICLR) 2025
GroupBind is a novel molecular docking framework that simultaneously considers multiple ligands docking to a protein.
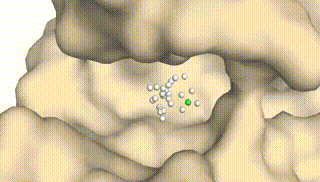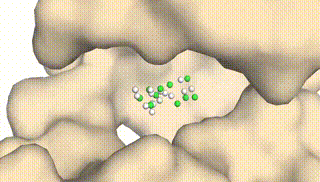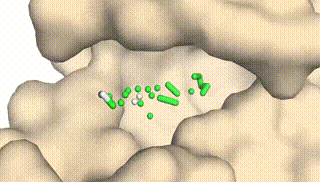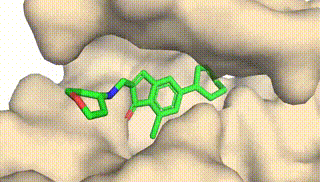
Integrating Protein Dynamics into Structure-Based Drug Design via Full-Atom Stochastic Flows
Xiangxin Zhou*, Yi Xiao*, Haowei Lin, Xinheng He, Jiaqi Guan, Yang Wang, Qiang Liu, Feng Zhou, Liang Wang, Jianzhu Ma (* equal contribution)
International Conference on Learning Representations (ICLR) 2025
DynamicFlow is a full-atom (stochastic) flow model that learns to transform apo pockets and noisy ligands into holo pockets and corresponding 3D ligand molecules.
Integrating Protein Dynamics into Structure-Based Drug Design via Full-Atom Stochastic Flows
Xiangxin Zhou*, Yi Xiao*, Haowei Lin, Xinheng He, Jiaqi Guan, Yang Wang, Qiang Liu, Feng Zhou, Liang Wang, Jianzhu Ma (* equal contribution)
International Conference on Learning Representations (ICLR) 2025
DynamicFlow is a full-atom (stochastic) flow model that learns to transform apo pockets and noisy ligands into holo pockets and corresponding 3D ligand molecules.
2024

Binding-Adaptive Diffusion Models for Structure-Based Drug Design
Zhilin Huang*, Ling Yang*, Zaixi Zhang, Xiangxin Zhou, Yu Bao, Xiawu Zheng, Yuwei Yang, Yu Wang, Wenming Yang (* equal contribution)
AAAI Conference on Artificial Intelligence (AAAI) 2024
BindDM extracts subcomplex from protein-ligand complex, and utilizes it to enhance the binding-adaptive 3D molecule generation.
Binding-Adaptive Diffusion Models for Structure-Based Drug Design
Zhilin Huang*, Ling Yang*, Zaixi Zhang, Xiangxin Zhou, Yu Bao, Xiawu Zheng, Yuwei Yang, Yu Wang, Wenming Yang (* equal contribution)
AAAI Conference on Artificial Intelligence (AAAI) 2024
BindDM extracts subcomplex from protein-ligand complex, and utilizes it to enhance the binding-adaptive 3D molecule generation.
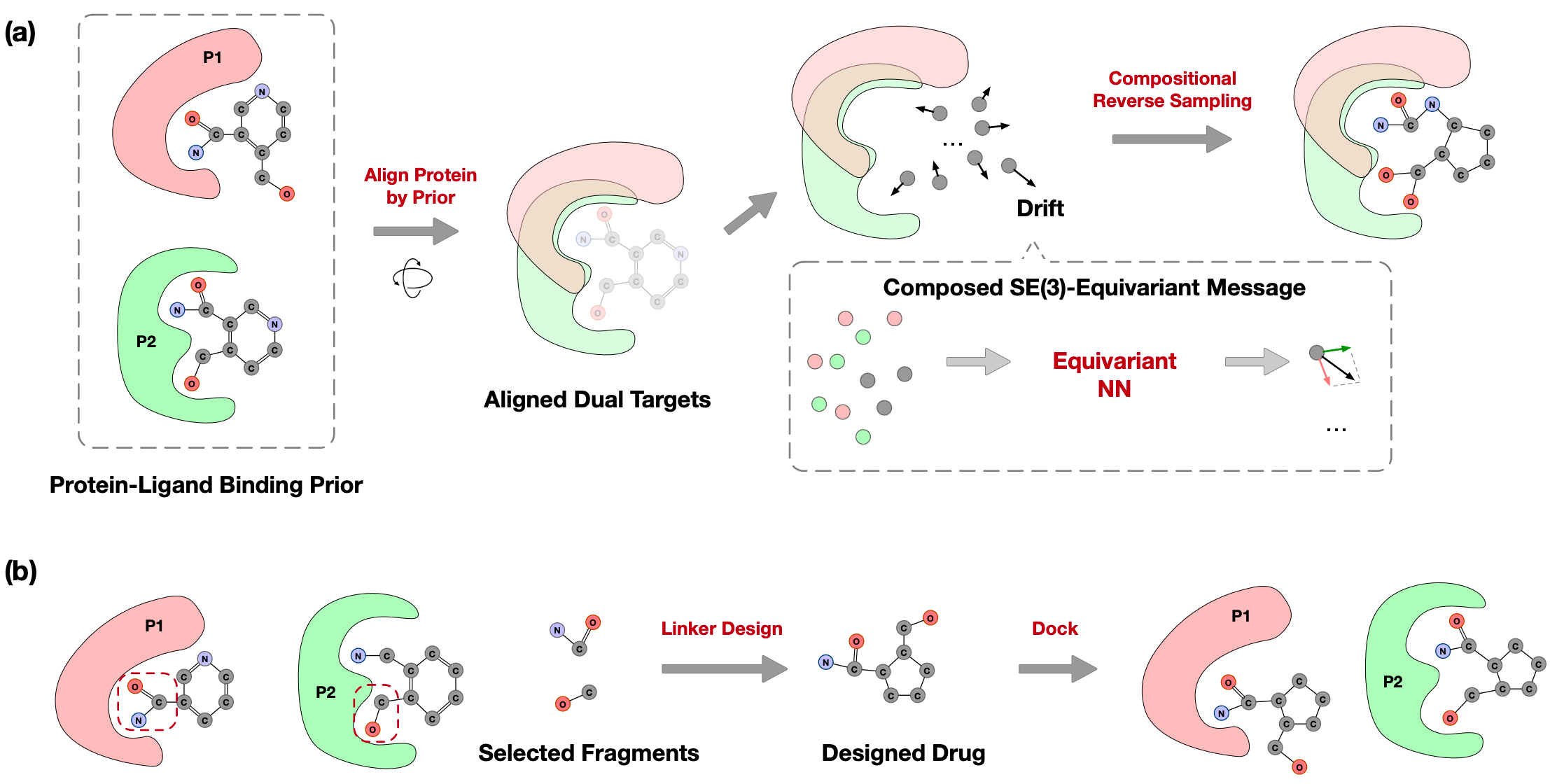
Reprogramming Pretrained Target-Specific Diffusion Models for Dual-Target Drug Design
Xiangxin Zhou, Jiaqi Guan, Yijia Zhang, Xingang Peng, Liang Wang, Jianzhu Ma
Conference on Neural Information Processing Systems (NeurIPS) 2024
DualDiff generates dual-target ligand molecules via compositional sampling based on single-target diffusion models.
Reprogramming Pretrained Target-Specific Diffusion Models for Dual-Target Drug Design
Xiangxin Zhou, Jiaqi Guan, Yijia Zhang, Xingang Peng, Liang Wang, Jianzhu Ma
Conference on Neural Information Processing Systems (NeurIPS) 2024
DualDiff generates dual-target ligand molecules via compositional sampling based on single-target diffusion models.
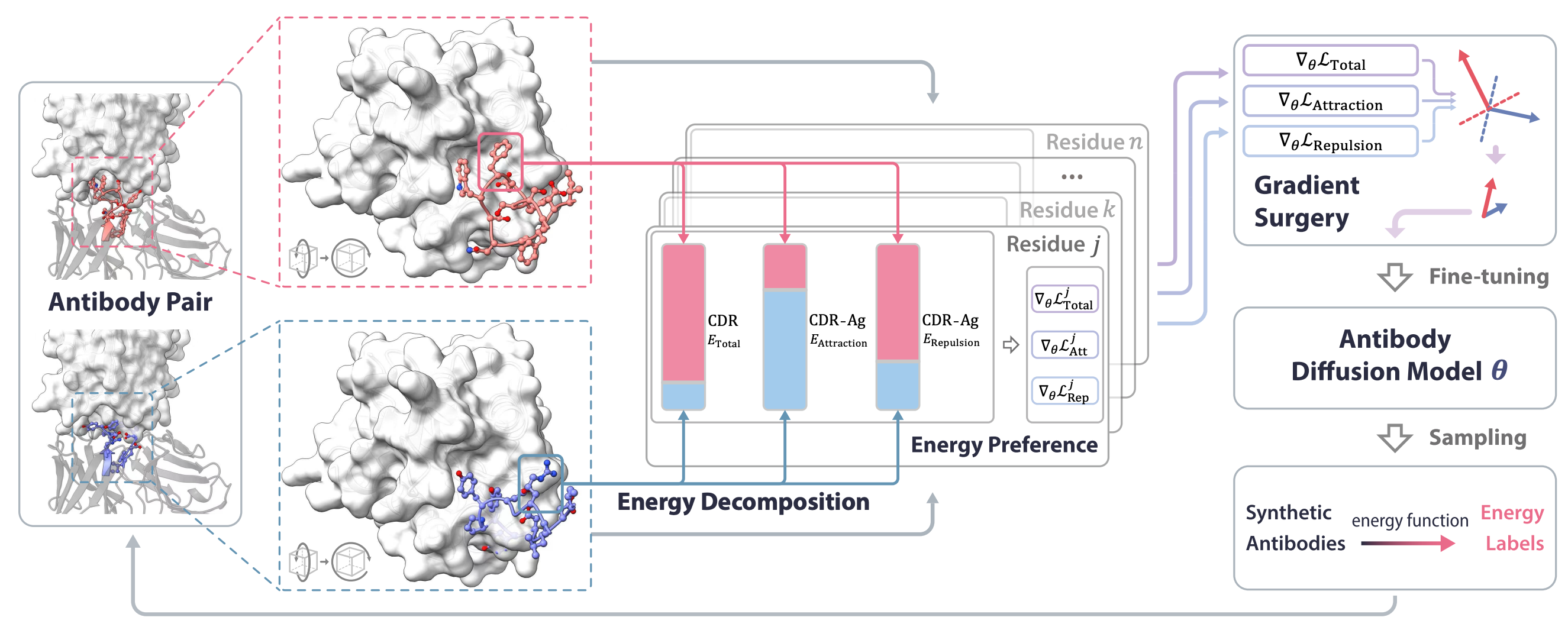
Antigen-Specific Antibody Design via Direct Energy-based Preference Optimization
Xiangxin Zhou*, Dongyu Xue*, Ruizhe Chen*, Zaixiang Zheng, Liang Wang, Quanquan Gu (* equal contribution)
Conference on Neural Information Processing Systems (NeurIPS) 2024
Direct energy-based preference optimzation guides the generation of antibodies with both rational structures and considerable binding affinities to given antigens.
Antigen-Specific Antibody Design via Direct Energy-based Preference Optimization
Xiangxin Zhou*, Dongyu Xue*, Ruizhe Chen*, Zaixiang Zheng, Liang Wang, Quanquan Gu (* equal contribution)
Conference on Neural Information Processing Systems (NeurIPS) 2024
Direct energy-based preference optimzation guides the generation of antibodies with both rational structures and considerable binding affinities to given antigens.
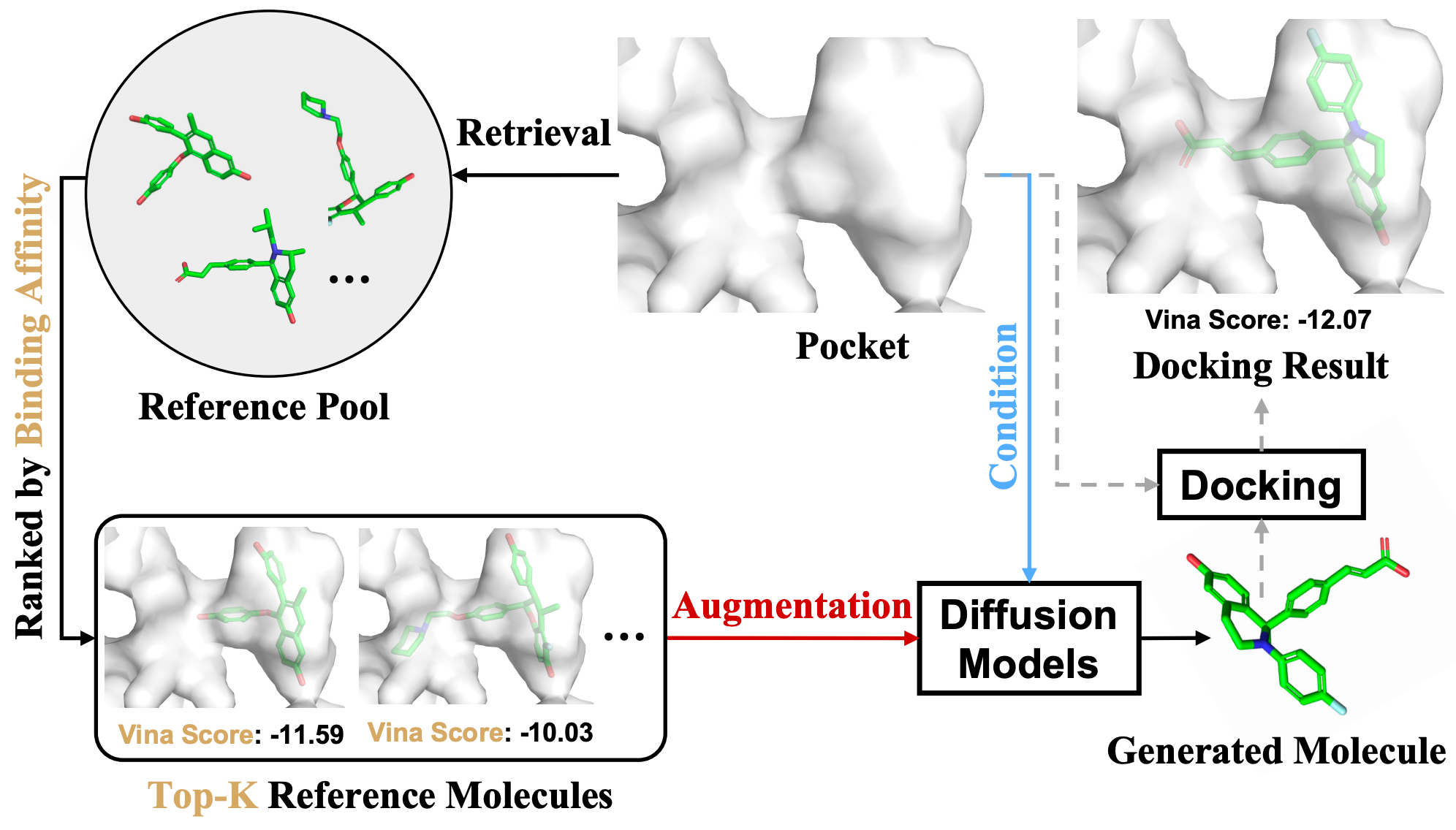
Interaction-based Retrieval-augmented Diffusion Models for Protein-specific 3D Molecule Generation
Zhilin Huang*, Ling Yang*, Xiangxin Zhou, Chujun Qin, Yijie Yu, Xiawu Zheng, Zikun Zhou, Wentao Zhang, Yu Wang, Wenming Yang (* equal contribution)
International Conference on Machine Learning (ICML) 2024
IRDiff leverages a curated set of ligand references, i.e., those with desired properties such as high binding affinity, to steer the diffusion model towards synthesizing ligands that satisfy design criteria.
Interaction-based Retrieval-augmented Diffusion Models for Protein-specific 3D Molecule Generation
Zhilin Huang*, Ling Yang*, Xiangxin Zhou, Chujun Qin, Yijie Yu, Xiawu Zheng, Zikun Zhou, Wentao Zhang, Yu Wang, Wenming Yang (* equal contribution)
International Conference on Machine Learning (ICML) 2024
IRDiff leverages a curated set of ligand references, i.e., those with desired properties such as high binding affinity, to steer the diffusion model towards synthesizing ligands that satisfy design criteria.
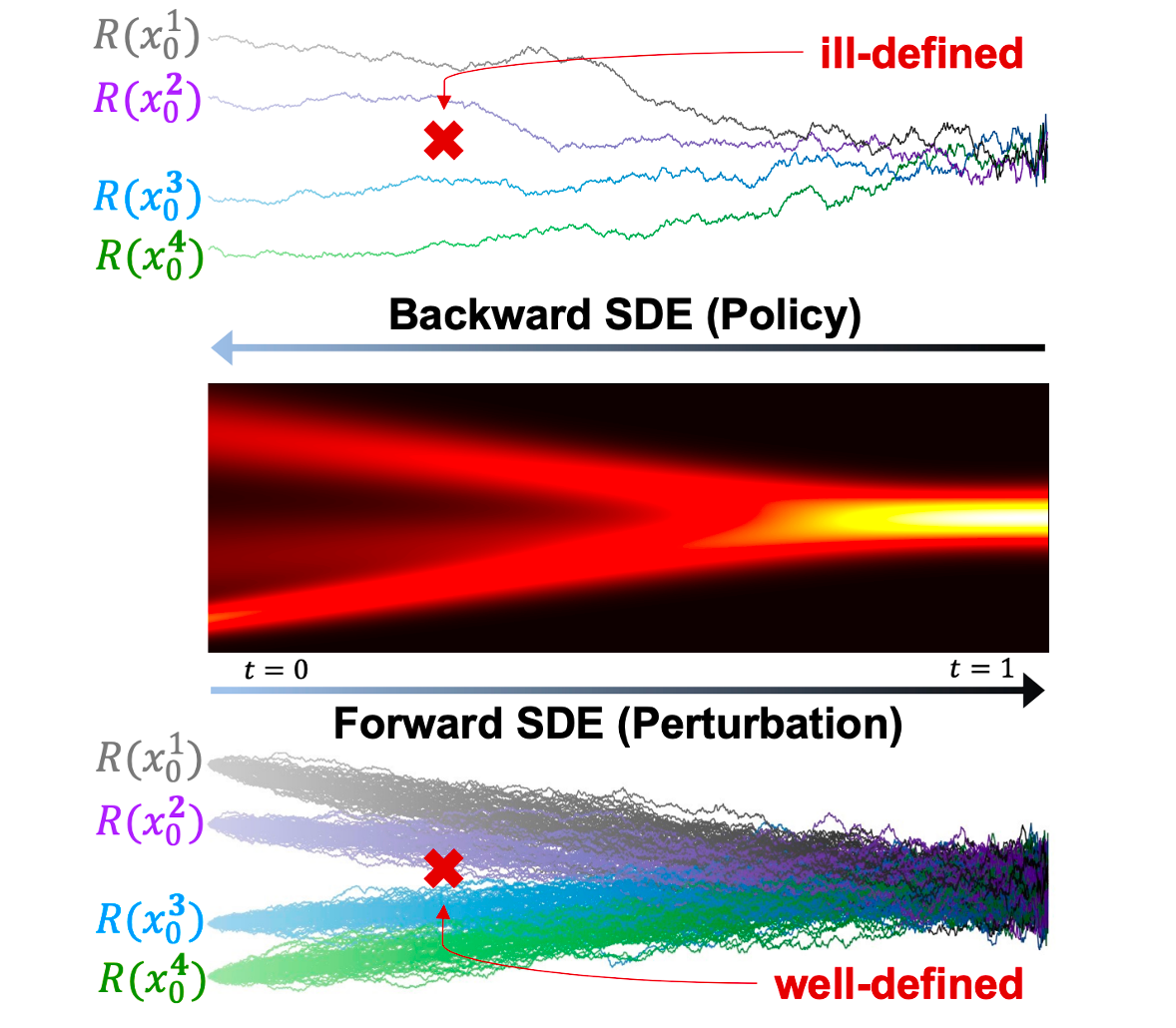
Stabilizing Policy Gradients for Stochastic Differential Equations via Consistency with Perturbation Process
Xiangxin Zhou, Liang Wang, Yichi Zhou
International Conference on Machine Learning (ICML) 2024
Policy gradients in data-scarce regions are ill-defined, leading to instability. Consistency ensured via score matching allows us to correctly estimate the policy gradients with sufficient data that can be efficiently sampled from the forward SDE (i.e., perturbation).
Stabilizing Policy Gradients for Stochastic Differential Equations via Consistency with Perturbation Process
Xiangxin Zhou, Liang Wang, Yichi Zhou
International Conference on Machine Learning (ICML) 2024
Policy gradients in data-scarce regions are ill-defined, leading to instability. Consistency ensured via score matching allows us to correctly estimate the policy gradients with sufficient data that can be efficiently sampled from the forward SDE (i.e., perturbation).

Protein-Ligand Interaction Prior for Binding-aware 3D Molecule Diffusion Models
Zhilin Huang*, Ling Yang*, Xiangxin Zhou, Zhilong Zhang, Wentao Zhang, Xiawu Zheng, Jie Chen, Yu Wang, Bin Cui, Wenming Yang (* equal contribution)
International Conference on Learning Representations (ICLR) 2024
Interaction Prior-guided Diffusion model (IPDiff) introduces geometric protein-ligand interactions into both diffusion and sampling process for the protein-specific 3D molecular generation.
Protein-Ligand Interaction Prior for Binding-aware 3D Molecule Diffusion Models
Zhilin Huang*, Ling Yang*, Xiangxin Zhou, Zhilong Zhang, Wentao Zhang, Xiawu Zheng, Jie Chen, Yu Wang, Bin Cui, Wenming Yang (* equal contribution)
International Conference on Learning Representations (ICLR) 2024
Interaction Prior-guided Diffusion model (IPDiff) introduces geometric protein-ligand interactions into both diffusion and sampling process for the protein-specific 3D molecular generation.
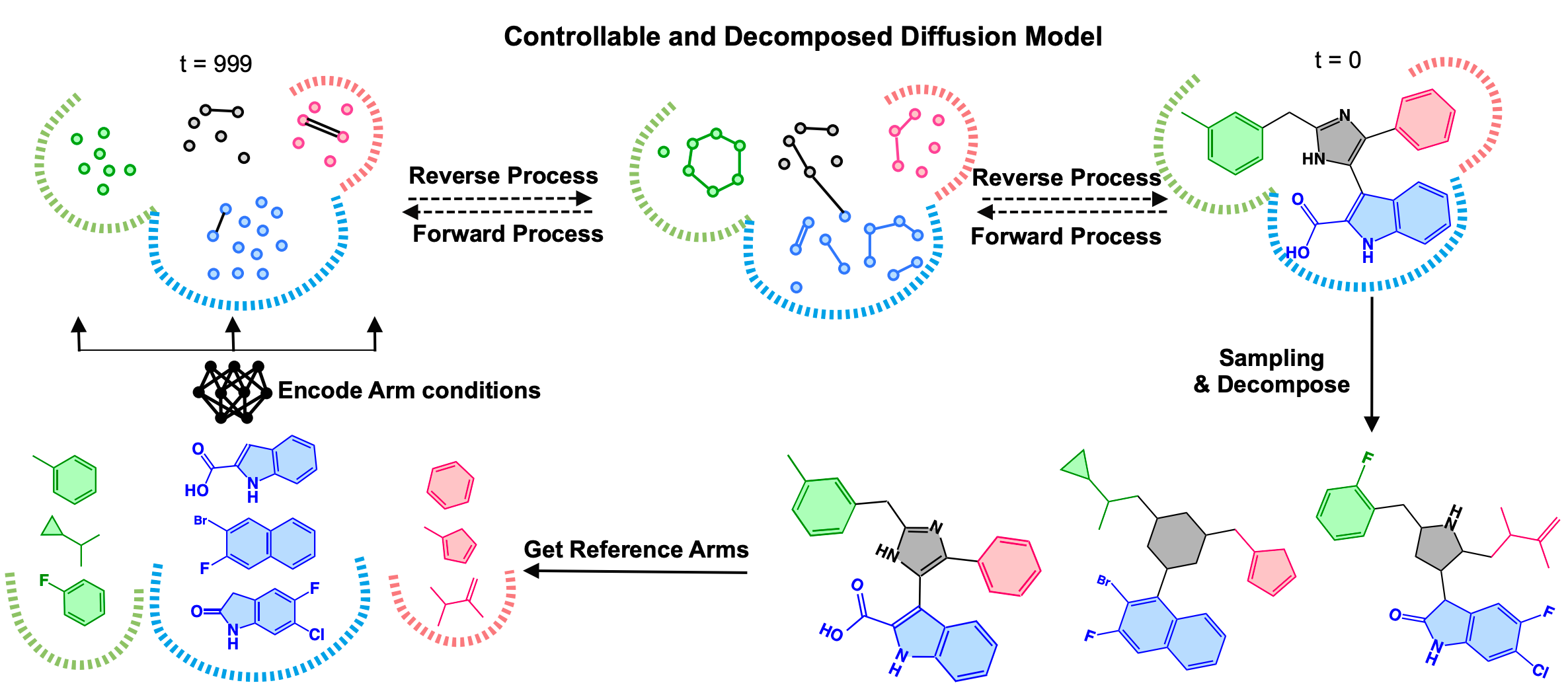
Controllable and Decomposed Diffusion Models for Structure-based Molecular Optimization
Xiangxin Zhou*, Xiwei Cheng*, Yuwei Yang, Yu Bao, Liang Wang, Quanquan Gu (* equal contribution)
International Conference on Learning Representations (ICLR) 2024
DecompOpt a structure-based molecular optimization method based on a controllable and decomposed diffusion model.
Controllable and Decomposed Diffusion Models for Structure-based Molecular Optimization
Xiangxin Zhou*, Xiwei Cheng*, Yuwei Yang, Yu Bao, Liang Wang, Quanquan Gu (* equal contribution)
International Conference on Learning Representations (ICLR) 2024
DecompOpt a structure-based molecular optimization method based on a controllable and decomposed diffusion model.
2023
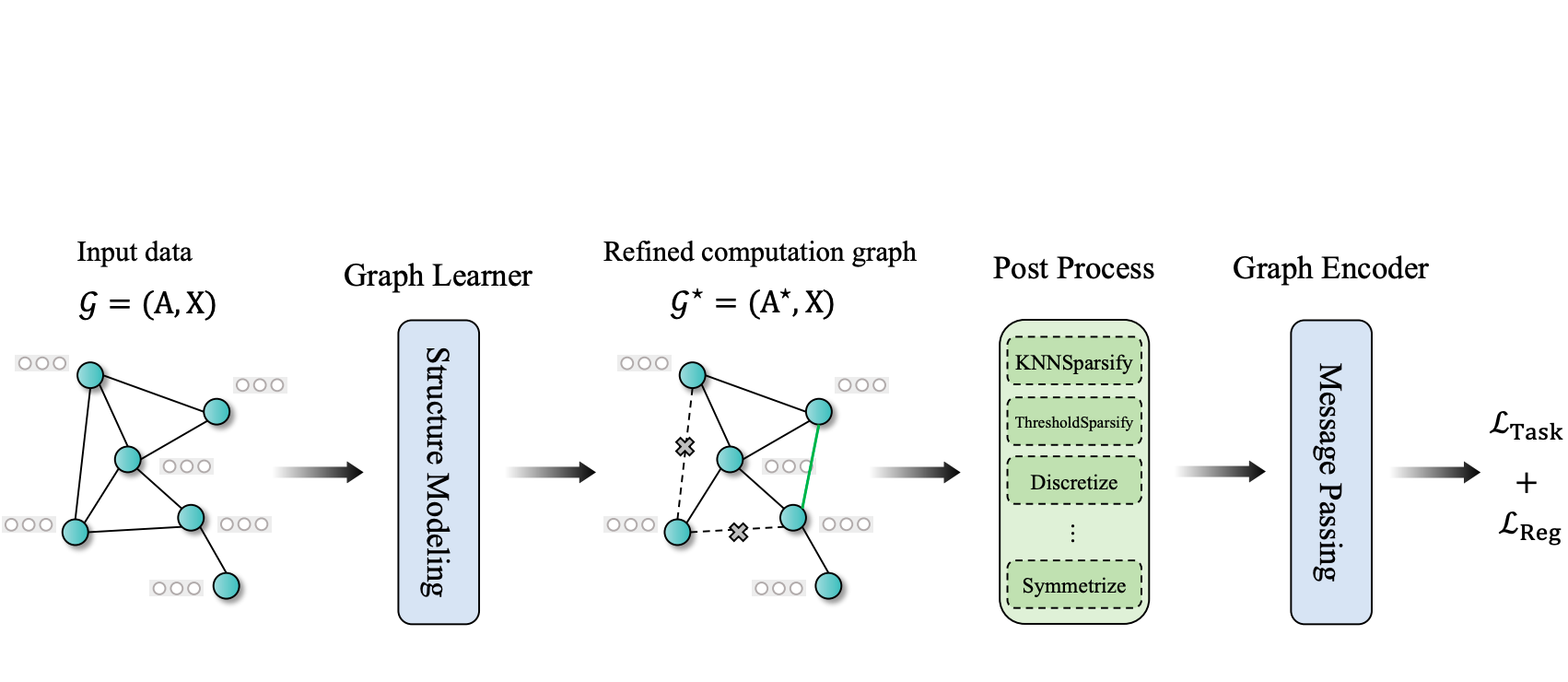
GSLB: The Graph Structure Learning Benchmark
Zhixun Li, Liang Wang, Xin Sun, Yifan Luo, Yanqiao Zhu, Dingshuo Chen, Yingtao Luo, Xiangxin Zhou, Qiang Liu, Shu Wu, Liang Wang, Jeffrey Xu Yu
Conference on Neural Information Processing Systems (NeurIPS) 2023
A comprehensive Graph Structure Learning Benchmark (GSLB) curated from 20 diverse graph datasets and 16 distinct GSL algorithms.
GSLB: The Graph Structure Learning Benchmark
Zhixun Li, Liang Wang, Xin Sun, Yifan Luo, Yanqiao Zhu, Dingshuo Chen, Yingtao Luo, Xiangxin Zhou, Qiang Liu, Shu Wu, Liang Wang, Jeffrey Xu Yu
Conference on Neural Information Processing Systems (NeurIPS) 2023
A comprehensive Graph Structure Learning Benchmark (GSLB) curated from 20 diverse graph datasets and 16 distinct GSL algorithms.
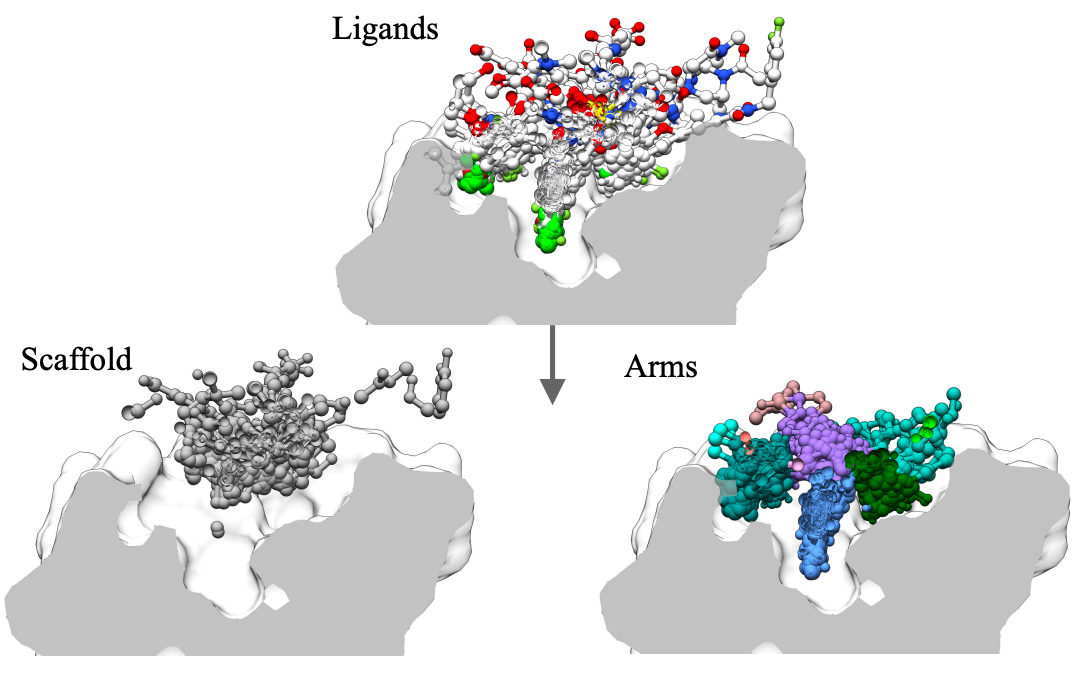
DecompDiff: Diffusion Models with Decomposed Priors for Structure-Based Drug Design
Jiaqi Guan*, Xiangxin Zhou*#, Yuwei Yang, Yu Bao, Jian Peng, Jianzhu Ma, Qiang Liu, Liang Wang, Quanquan Gu# (* equal contribution, # corresponding author)
International Conference on Machine Learning (ICML) 2023
DecompDiff is a diffusion model for SBDD with decomposed priors over arms and scaffold, equipped with bond diffusion and additional validity guidance.
DecompDiff: Diffusion Models with Decomposed Priors for Structure-Based Drug Design
Jiaqi Guan*, Xiangxin Zhou*#, Yuwei Yang, Yu Bao, Jian Peng, Jianzhu Ma, Qiang Liu, Liang Wang, Quanquan Gu# (* equal contribution, # corresponding author)
International Conference on Machine Learning (ICML) 2023
DecompDiff is a diffusion model for SBDD with decomposed priors over arms and scaffold, equipped with bond diffusion and additional validity guidance.
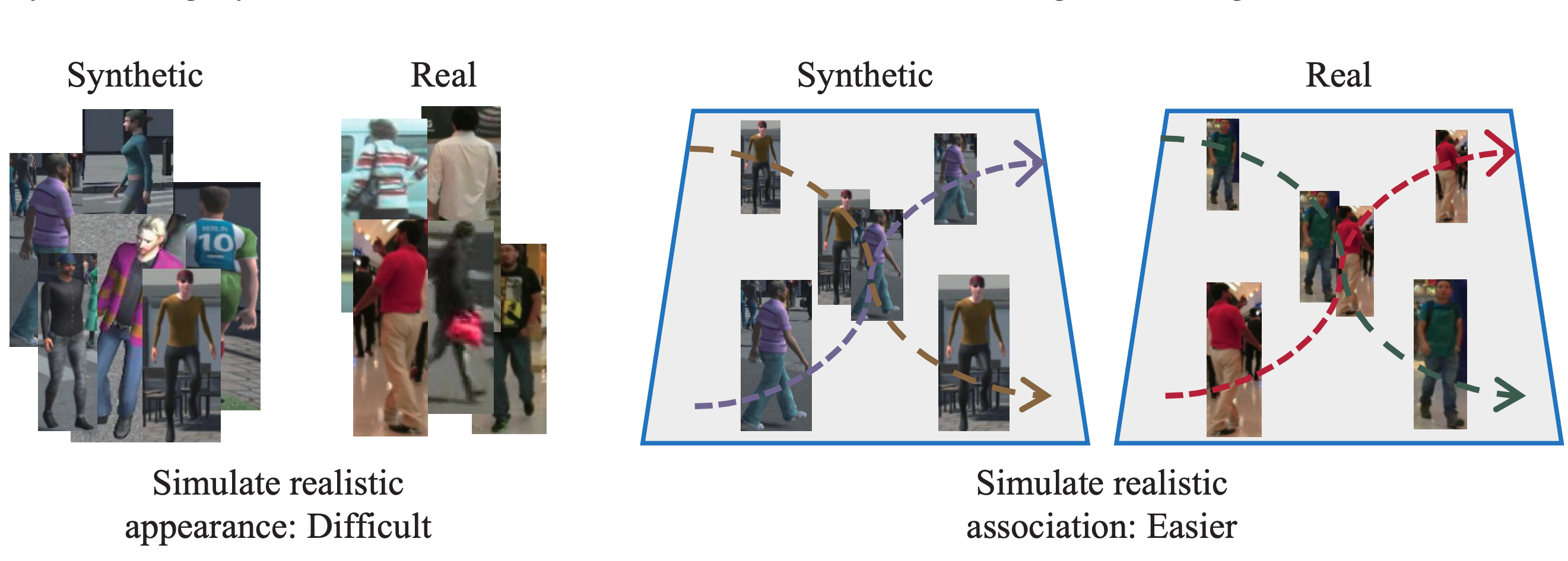
A Study of Using Synthetic Data for Effective Association Knowledge Learning
Yuchi Liu, Zhongdao Wang, Xiangxin Zhou, Liang Zheng
Machine Intelligence Research (MIR) 2023
This paper studies the role of synthetic data in multi-object tracking.
A Study of Using Synthetic Data for Effective Association Knowledge Learning
Yuchi Liu, Zhongdao Wang, Xiangxin Zhou, Liang Zheng
Machine Intelligence Research (MIR) 2023
This paper studies the role of synthetic data in multi-object tracking.
2021
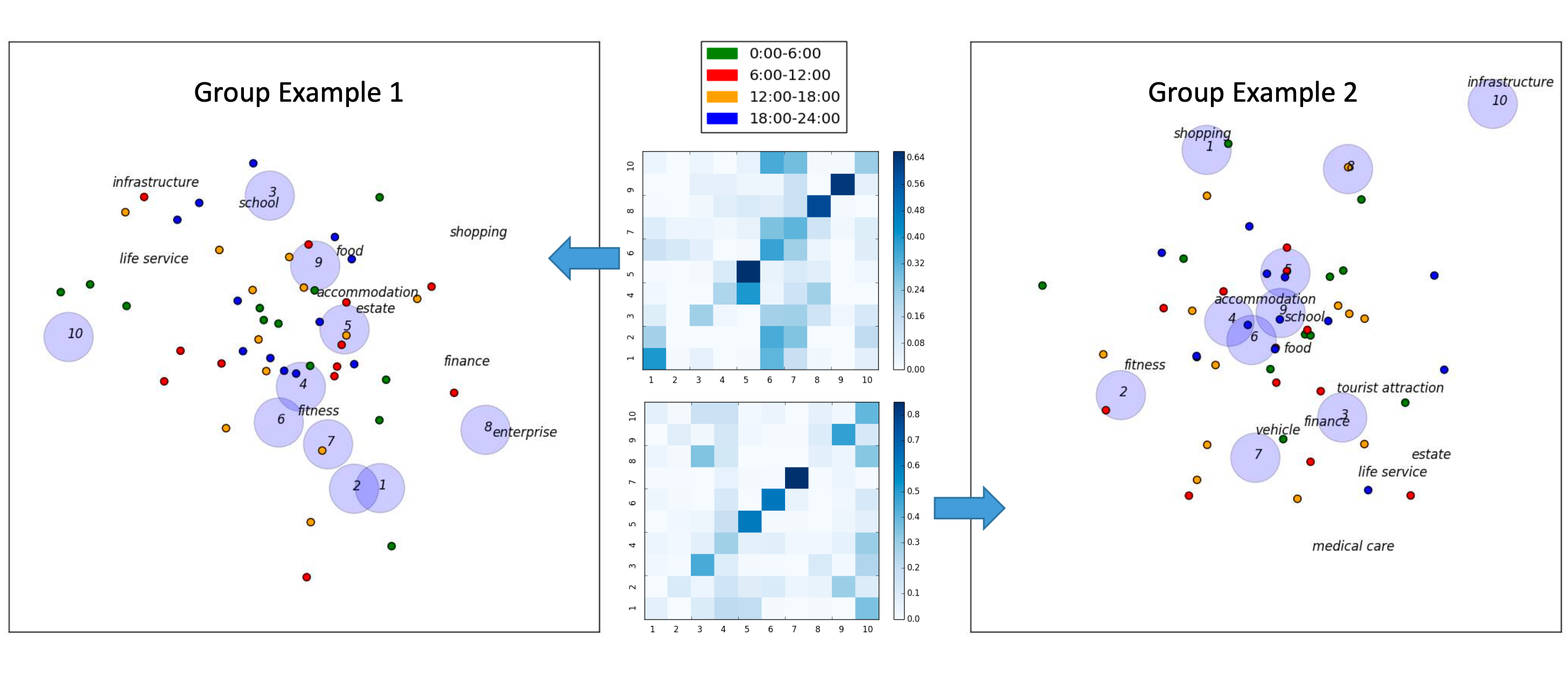
Semantics-Aware Hidden Markov Model for Human Mobility
Hongzhi Shi, Yong Li, Hancheng Cao, Xiangxin Zhou, Chao Zhang, Vassilis Kostakos
IEEE Transactions on Knowledge and Data Engineering (TKDE) 2021 Shorter version got accepted by SIAM International Conference on Data Mining (SDM), Full paper
A semantics-aware hidden Markov model for human mobility modeling that takes into account location, time, activity and user motivation behind human mobility as a whole.
Semantics-Aware Hidden Markov Model for Human Mobility
Hongzhi Shi, Yong Li, Hancheng Cao, Xiangxin Zhou, Chao Zhang, Vassilis Kostakos
IEEE Transactions on Knowledge and Data Engineering (TKDE) 2021 Shorter version got accepted by SIAM International Conference on Data Mining (SDM), Full paper
A semantics-aware hidden Markov model for human mobility modeling that takes into account location, time, activity and user motivation behind human mobility as a whole.
2019

Global Sparse Momentum SGD for Pruning Very Deep Neural Networks
Xiaohan Ding, Guiguang Ding, Xiangxin Zhou, Yuchen Guo, Jungong Han, Ji Liu
Conference on Neural Information Processing Systems (NeurIPS) 2019
A novel momentum-SGD-based optimization method that reduces the network complexity by on-the-fly pruning.
Global Sparse Momentum SGD for Pruning Very Deep Neural Networks
Xiaohan Ding, Guiguang Ding, Xiangxin Zhou, Yuchen Guo, Jungong Han, Ji Liu
Conference on Neural Information Processing Systems (NeurIPS) 2019
A novel momentum-SGD-based optimization method that reduces the network complexity by on-the-fly pruning.

Class-balanced Grouping and Sampling for Point Cloud 3D Object Detection
Benjin Zhu, Zhengkai Jiang, Xiangxin Zhou, Zeming Li, Gang Yu
Technical Report 2019 Winner of CVPR 2019 WAD NuScenes Challenge
Our 3D object detection solution: Sparse 3D Convolution feature extractor + Region Proposal Network + class-balanced multi-head network.
Class-balanced Grouping and Sampling for Point Cloud 3D Object Detection
Benjin Zhu, Zhengkai Jiang, Xiangxin Zhou, Zeming Li, Gang Yu
Technical Report 2019 Winner of CVPR 2019 WAD NuScenes Challenge
Our 3D object detection solution: Sparse 3D Convolution feature extractor + Region Proposal Network + class-balanced multi-head network.